nf-core/viralmetagenome

A bioinformatics pipeline for untargeted viral genome reconstruction and identification of intra-host variants from metagenomic sequencing data
nf-core/viralmetagenome is a comprehensive bioinformatics pipeline for untargeted viral genome reconstruction and intra-host variant identification from metagenomic sequencing data. Built with within nf-core, it follows best practices for reproducible and scalable bioinformatics workflows.
As the main developer, I created viralmetagenome to address the need for standardized viral genomics analysis in metagenomic datasets. The pipeline enables researchers to reconstruct high-quality viral genomes from complex samples and identify genetic variants crucial for understanding viral evolution, drug resistance, and pathogenesis.
Workflow overview:
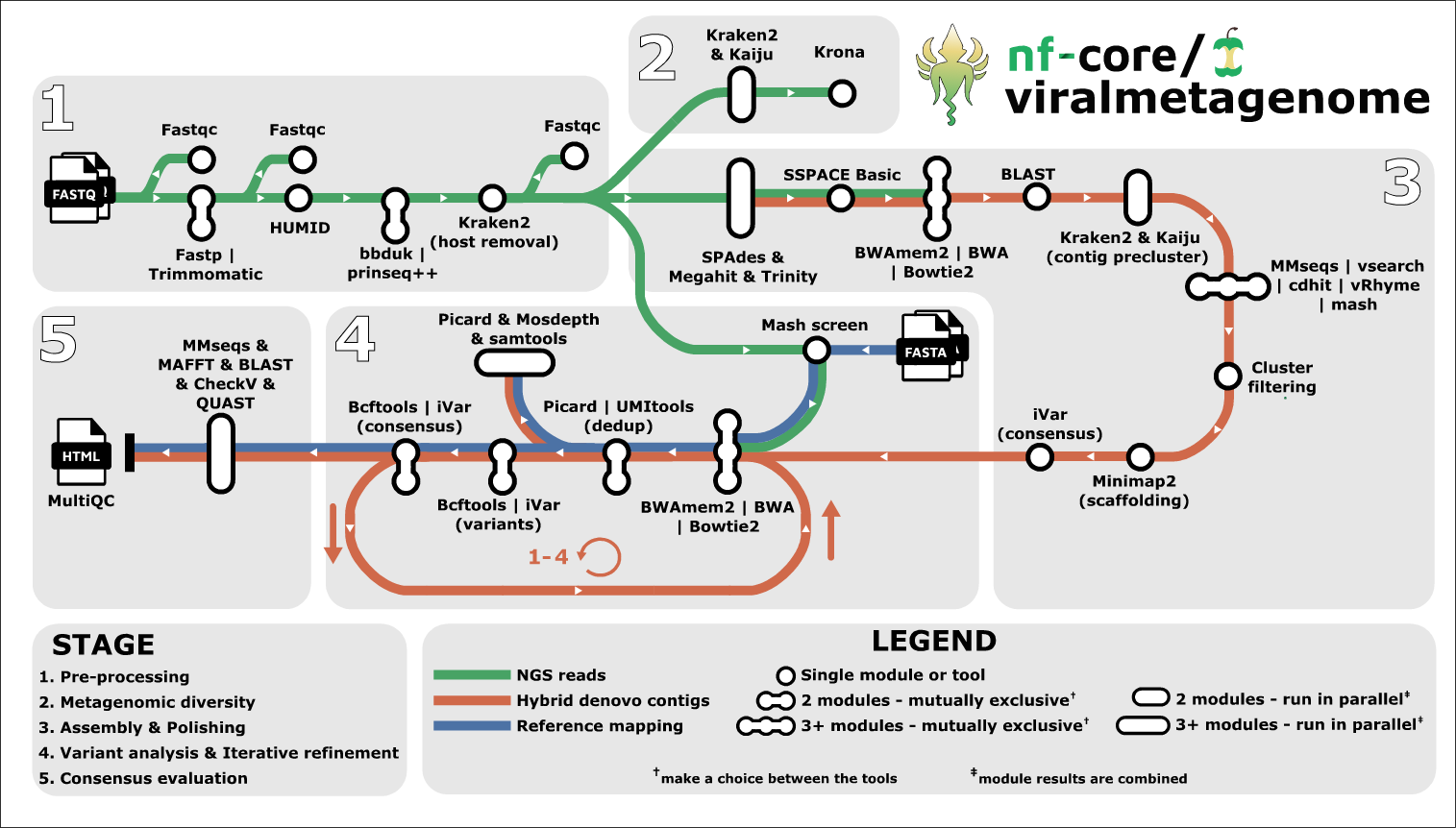
The complete source code, documentation, and usage examples are available on GitHub at nf-core/viralmetagenome.
Completed the migration from personal reposistory to nf-core and renamed the pipeline from viralgenie to viralmetagenome
First complete version of viralmetagenome
Started development of viralmetagenome under the name of viralgenie pipeline for own sequencing projects

