ggphylogeo

An R package for visualizing phylogenetic trees on geographical maps
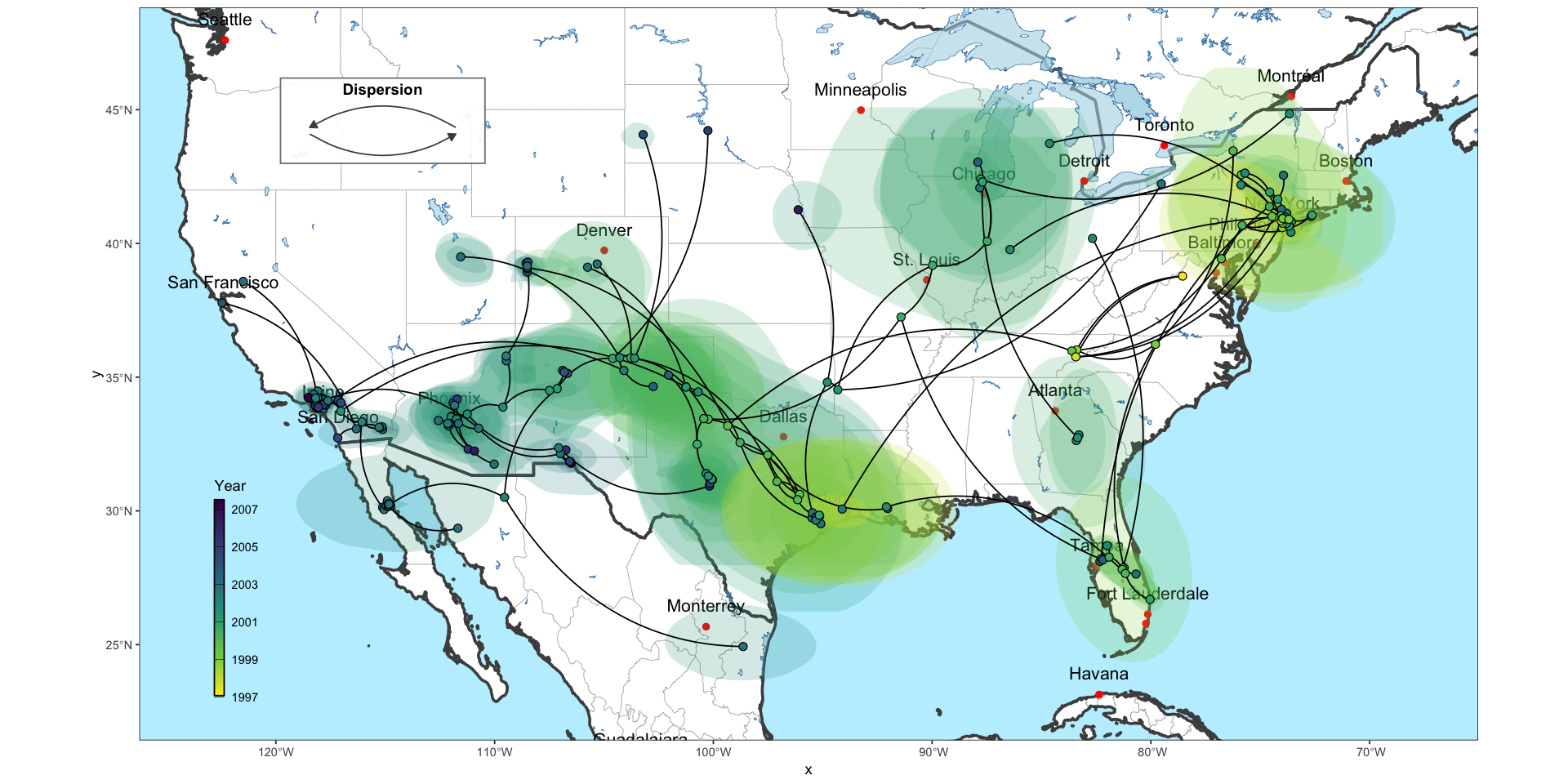
ggphylogeo is an R package designed to bridge the gap between phylogenetics and cartography. It allows researchers to project phylogenetic trees directly onto geographical maps, providing a clear visual representation of viral spread or species migration over space and time.
Visualizing phylogeographic data often requires juggling multiple complex libraries and various tools. I developed ggphylogeo to streamline this process, leveraging the power of ggplot2 to create publication-ready figures with minimal code.
Key features include:
- Geospatial projection: Seamless integration of tree topology with latitude/longitude coordinates.
- Customizable aesthetics: Full control over edge coloring, node sizing, and map layers.
The source code and vignettes for creating your own phylogeographic plots are available on GitHub.
